Lucie Bittner, PhD
Associate Professor
Bioinformatics, Environmental and Evolutionary Genomics
ML4Oceans summer School 2022 – announcement
The use of artificial intelligence is now crucial for environmental and climate issues, including oceanography. In this context, SCAI, ISCD and the Institut de l’Océan of Sorbonne Université joined their forces to offer a transdisciplinary summer school for PhD and post-doctoral students.
20 students will be selected according to their academic background, their research subject & their motivation to participate in this school.
The objective: to acquire knowledges and skills in AI applied to oceanography (marine biology – omics, imagery -, physics, modeling, etc.).
Additional information here.
Registration is open here until May 31st 2022.




Montpellier Omics days 2022, France
– January 26th 2022
Montpellier OMICS Days (MOD)’ conference is organised by the Bioinformatics Master’s students and students studying Statistics and Data Science at the Faculty of Sciences of the University of Montpellier, France.
The 10th edition was held online.The programme can be found in the Programme tab and the conferences are available in the Vidéos tab.
Lucie Bittner was invited as a main speaker, and gave the first conference.
6th International Marine Connectivity Conference, MNHN, Paris, France – December 6th-8th 2022
This international conference was organised by the iMarCo and the SEA-UNICORN initiatives. It aimed to promote the development of an international network for promoting collaborative projects among European scientists interested in the study of marine connectivity.
Lucie Bittner presented results from genetic differentiation studies of three pico-phytoplankton species, based on metagenomics data.

Environmental genomics congress, Tours, France – October 27th-29th 2021

Pavla Debeljak presented her investigations on the small green algae ‘dark matter’.
First meeting on site since winter 2019-2020 !
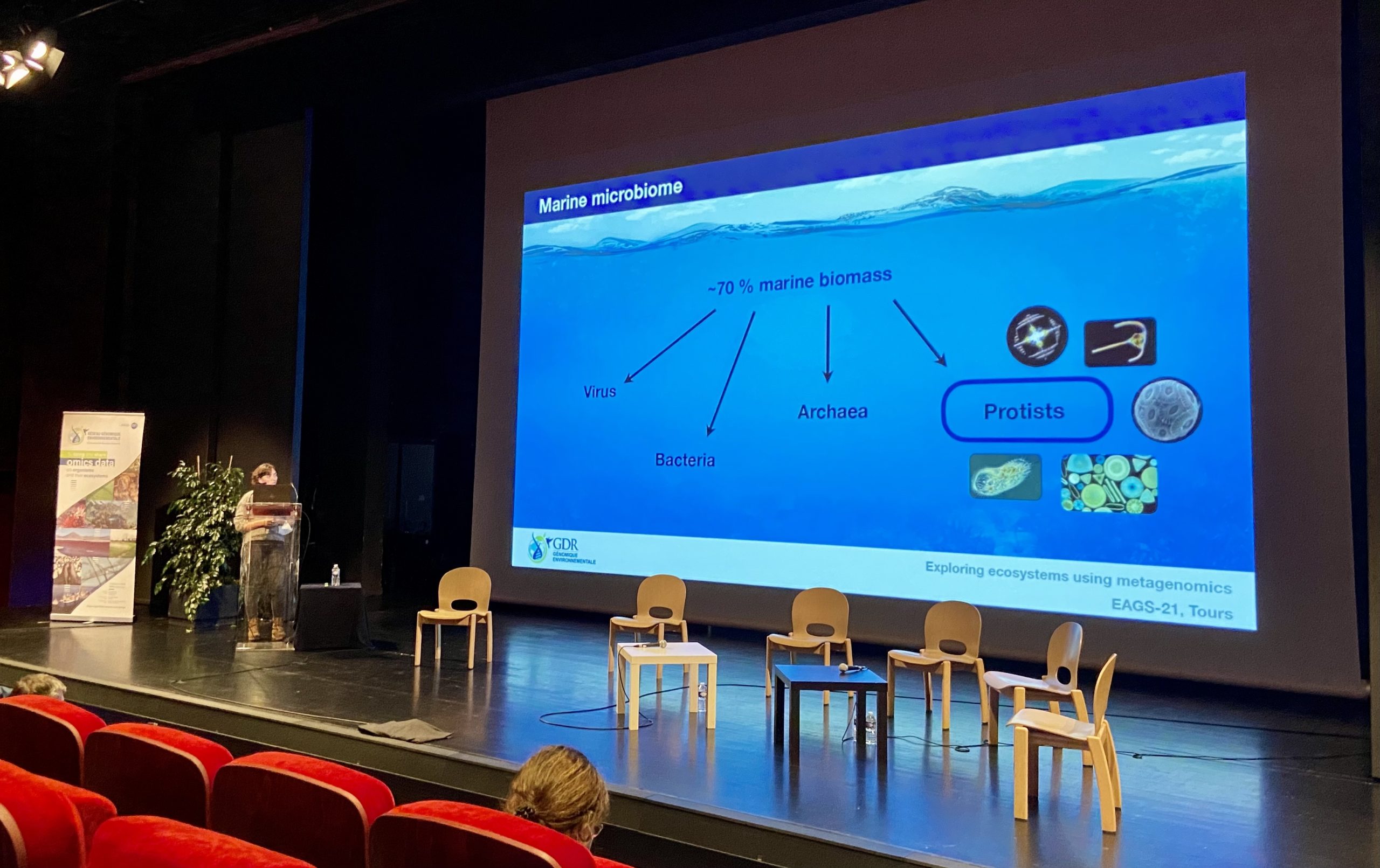
Iris Rizos showed her master2 results to expand our idea of taxonomic protist diversity.
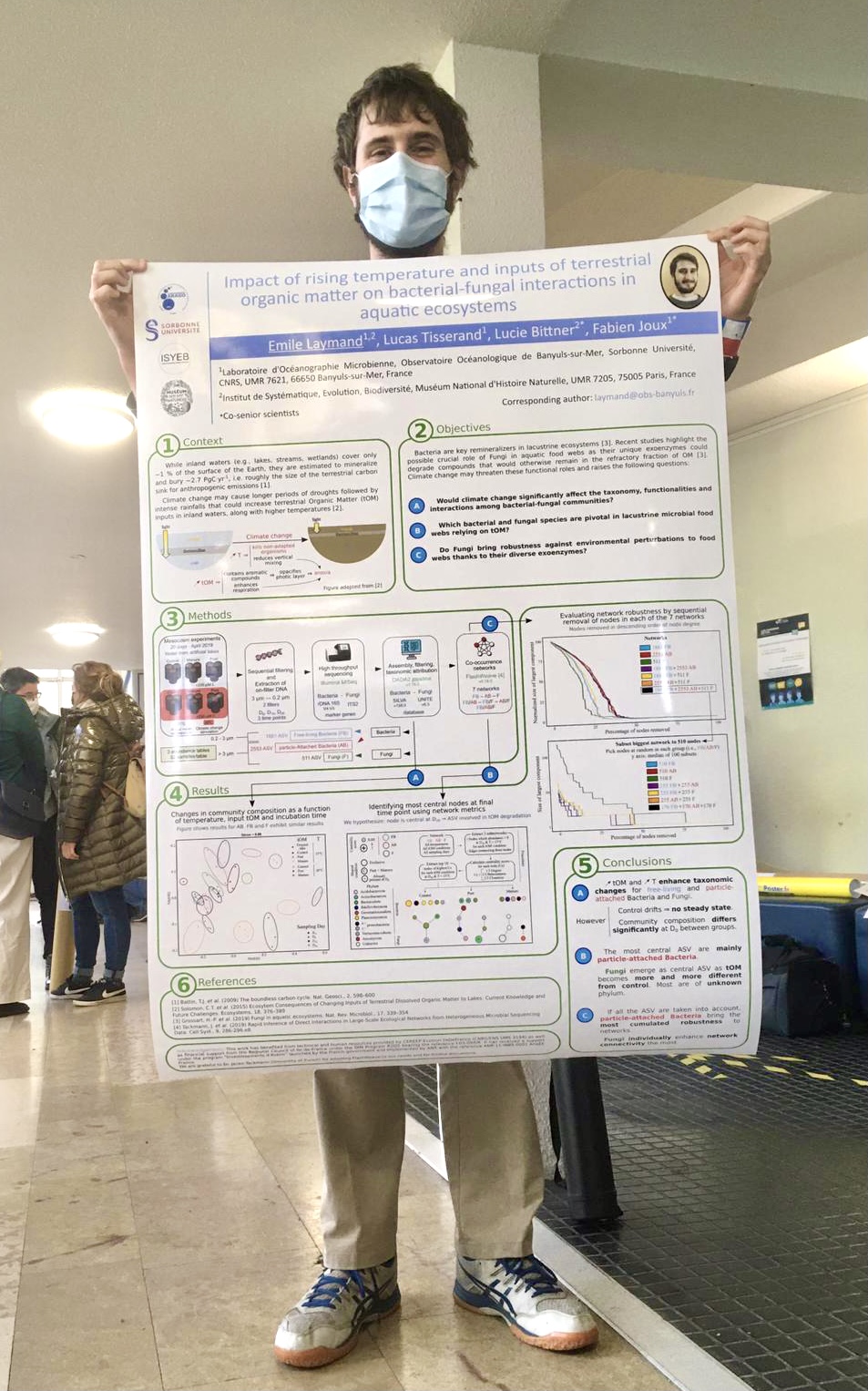
Emile Laymand discussed his master2 results: i.e., what would be the impact of rising temperature on bacterial-fungal interactions in aquatic ecosystems.
Ophelie Da Silva PhD’s defense – September 30th 2021
Structure of the planktonic ecosystem : contribution of high-throughput sequencing and imaging data
Her PhD manuscript is available here.
Keywords. Plankton, Biogeography, Meta-omics, Imaging, Environment.
Abstract: Planktonic organisms are key actors in oceanic ecosystems, which support trophic networks and play a major role in biogeochemical cycles and climate regulation. While the spatio-temporal distribution of planktonic diversity can be investigated at several levels, from the gene to the ecosystem, identifying the underlying mechanisms is challenging. Indeed, the structure of diversity results from different evolutionary and ecological processes that can act simultaneously. Since the beginning of the 21st century, the oceanic environment has been increasingly monitored. Numerous observation platforms have been deployed, leading to the acquisition of a large amount of data for multiple environmental characteristics. At the same time, technologies for studying living organisms have been developed. Thus, an unprecedented sampling of planktonic organisms has taken place. In particular, high-throughput sequencing and imaging data provide molecular, taxonomic and functional information at several biological levels. The objective of this thesis was to explore the structure of planktonic ecosystems using high-throughput sequencing and imaging data. Coupling with environmental data could contribute to a better understanding of the spatial distribution of planktonic diversity, from species to communities. In the first part, the genetic diversity of protists was studied at the species level. The hypothesis was that metagenomics could provide access to the poorly characterized spatial organization of the intraspecific protist genetic diversity, as well as to the mechanisms underlying it. In a second part, the link between genetic diversity and functional diversity was explored. Transparency was targeted. This functional trait is little explored at the community level and its molecular basis is poorly identified. A data-driven approach allowed this trait to emerge from imaging data, leading to the exploration of its biogeography and molecular basis. In the last part, the high potential of complementarity between sequencing, imaging and environmental datasets was explored, in order to highlight the multi-scale structure of the planktonic ecosystem and to identify its global structure. Finally, all the results were discussed to highlight the contributions that these data can provide to the understanding of planktonic ecosystems, as well as the limitations they can face.
Collaborations between the Laboratoire d’Océanographie de Villefranche (Villefranche-sur-mer) and the Institut de Systématique, Évolution, Biodiversité (Paris).



Emile Faure PhD’s defense – December 2nd 2020
Contributions of meta-omics data to the detection of functional traits in planktonic ecosystems
Keywords. Plankton, Omics, Biogeochemistry, Functional traits.
The defense is available on youtube. His manuscript is available here.
The jury was composed of:
- Sébastien Monchy, Professeur (Université du Littoral Côte d’Opale), Rapporteur
- Christian Tamburini, DR CNRS (Mediterranean Institute of Oceanography), Rapporteur
- Thomas Mock, Professor (University of East Anglia), Examinateur
- Ingrid Obernosterer, DR CNRS (Sorbonne Université), Examinatrice
- Sakina-Dorothée Ayata, MC (Sorbonne Université), Co-Directrice de thèse
- Lucie Bittner, MC (Sorbonne Université), Co-Directrice de thèse
Abstract: Marine planktonic organisms play a crucial role in trophic networks, global biogeochemical cycles, and climate regulation. Biogeochemical models simulate planktonic ecosystems dynamics to understand and predict climate change. In most biogeochemical models, planktonic diversity is implemented either through plankton functional types (PFT), i.e. theoretical entities grouping planktonic organisms according to shared functional capacities (e.g., calcifiers, nitrogen fixers or silicifiers), or functional traits, i.e. morphological, physiological or phenological features measurable at the individual level that effect growth, reproduction or survival (e.g. feeding modes, production of toxins or body size). These methods imply an a priori and restricted choice of the considered types or traits of planktonic organisms, potentially leading to oversimplified representations of planktonic diversity in models. Unprecedented amounts of meta-omics data on marine planktonic communities were recently collected at global scales, calling for the use of data-driven methodologies to determine and quantify the potential and realized functional traits of planktonic organisms in-situ. My objective in this thesis was therefore to determine how to use meta-omics data to quantify the distribution of functional traits in the environment. In a first part, I present how omics data can be used to describe and quantify specific, a priori selected traits in the global ocean. A particular focus is made on two functional traits: mixotrophy, from which the genomic basis is poorly known, and dimethyl sulfide (DMS) production, from which the genomic basis is well understood. I show how metabarcoding data on one hand and functional genomic markers on the other hand allow to decipher the biogeography of functional traits, identifying limits and advantages of the two types of data. In a second part, I present an approach allowing to detect putative protein families in metagenomics data that can be associated with functional traits, without any a priori choice of functional traits of interest. By quantifying the response of these emergent clusters to physico-chemical gradients in the global ocean, I show how this approach could allow to predict the functional composition of planktonic communities from environmental data in the near future. Finally, I use my results to discuss the potential of meta-omics data as a means of realistically representing the diversity of planktonic communities in biogeochemical models.
Collaborations between the Institut de Systématique, Évolution, Biodiversité (Paris) and the Laboratoire d’Océanographie de Villefranche (Villefranche-sur-mer)



Pavla Debeljak has joined the lab ! – September 15th, 2020

As a post-doc, she will study the functional and taxonomical, unknown omics from the planktonic ecosystems.
She is founded by the H2020 Blue-Cloud project, and will contribute to the Plankton Genomics demonstrator.
Collaborations between the Institut de Systématique, Évolution, Biodiversité (Paris), the LAGE team at the CEA Genoscope (Evry) and the Laboratoire d’Océanographie de Villefranche (Villefranche-sur-mer)


 .
. 
Master 2 internship [ January 2021 – June 2021 ] – September 2020
Dynamics of aquatic microbial interactions involved in the degradation of terrestrial organic matter
Detailed description here
Keywords: co-occurrence networks, interactions, metabarcoding, bioinformatics/biostatistics, microbial ecology, organic matter degradation, temperature
Collaborations between the Institut de Systématique, Évolution, Biodiversité (Paris) and the Laboratoire d’Océanographie Microbienne (Banyuls-sur-mer)
Contacts: lucie.bittner@upmc.fr and joux@obs-banyuls.fr




